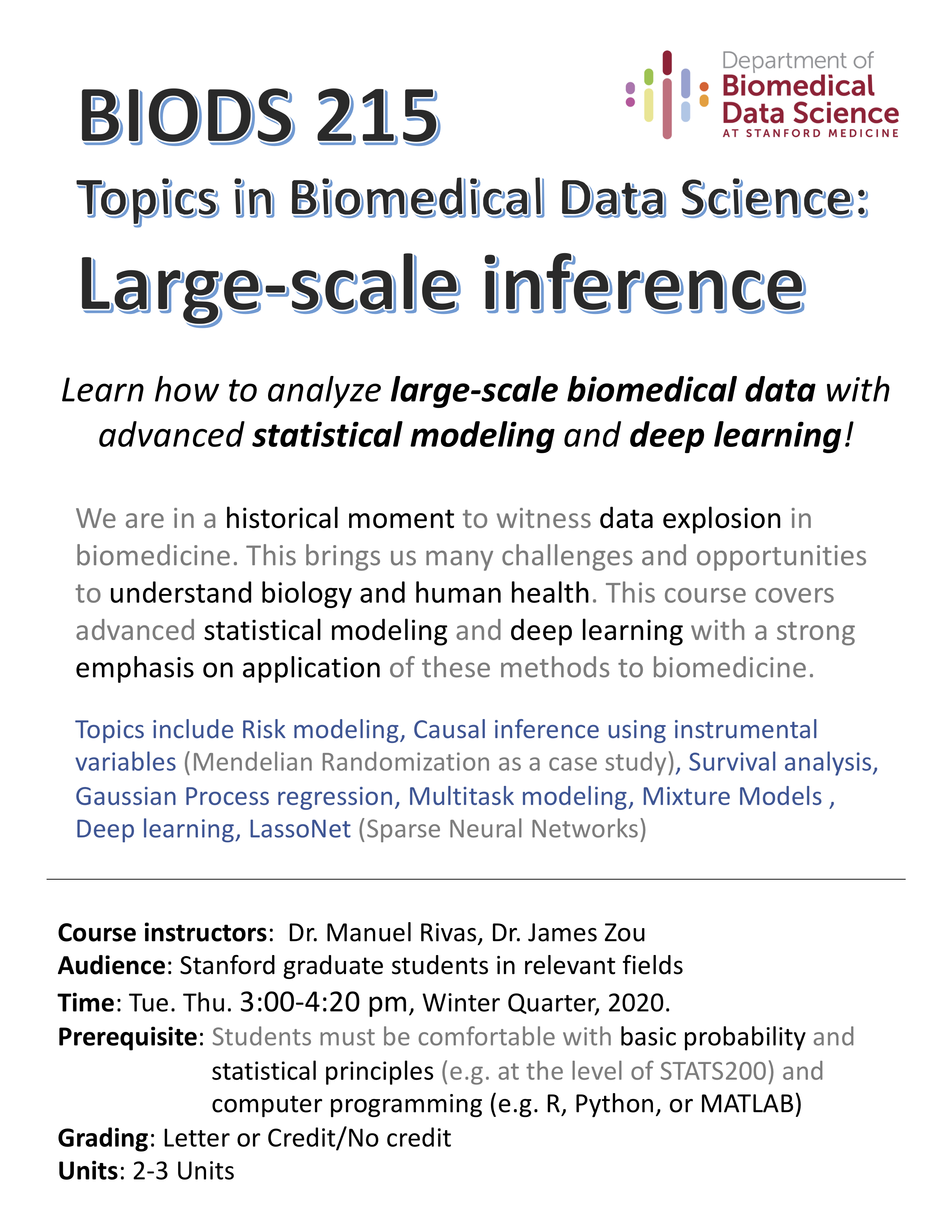
2020 Winter: BIODS 215
Welcome to BIODS215 Topics in Biomedical Data Science: Large-scale inference!
- Website for 2018 Winter quarter is available here
- Website for 2017 Spring quarter is available here
- Go back to the top page
Teaching team
We highly encourage everyone to use Piazza to contact with the teaching team.
However, you can also reach out to us using email.
The mailing list to the entire teaching team is: biods215-2020 at lists.stanford.edu.
Instructors
- Manuel A. Rivas (email:
mrivas at stanford.edu), Office hours: Tuesdays 4:20 - 5:20 (MSOB x321) - James Zou (email:
jamesz at stanford.edu)
TA
- Yosuke Tanigawa (email:
ytanigaw at stanford.edu), Office hours: Tuesdays 11:00 - 12:00 (MSOB x391)
Links
- Canvas
- Piazza
- Gradescope, Entry Code:MZGP3Y
Course content plan
| # | Date | Lecturer | Room | Topic |
|---|---|---|---|---|
| 1 | 1/7/2020 | Manuel | MSOBx303 | Overview of emerging topics in biomedical data science |
| 2 | 1/9/2020 | Manuel | MSOBx303 | Topics in Linear Algebra |
| 3 | 1/14/2020 | Manuel | MSOBx303 | Optimization |
| 4 | 1/16/2020 | Manuel | MSOBx303 | Mixture Models |
| 5 | 1/21/2020 | James | MSOBx303 | Machine Learning for CRISPR editing |
| 6 | 1/23/2020 | Guest lecture | MSOBx303 | Ismael Lemhadri: “lassoNet” |
| 7 | 1/28/2020 | Manuel | MSOBx303 | Causal inference using instrumental variables |
| 8 | 1/30/2020 | Manuel | MSOBx303 | Causal inference using instrumental variables II: Mendelian randomization |
| 9 | 2/4/2020 | Manuel | MSOBx303 | Gaussian Process regression |
| 10 | 2/6/2020 | Yosuke | MSOBx303 | Reproducible large-scale inference |
| 11 | 2/11/2020 | James | MSOBx303 | Deep Learning I |
| 12 | 2/13/2020 | James | LK120 | Deep Learning II |
| 13 | 2/18/2020 | Manuel | MSOBx303 | Risk Models |
| 14 | 2/20/2020 | Manuel | MSOBx303 | Survival risk models |
| 15 | 2/25/2020 | Manuel | MSOBx303 | Multitask risk modeling |
| 16 | 2/27/2020 | Manuel | MSOBx393 | False discovery rates |
| 17 | 3/3/2020 | Final project | MSOBx303 | Final project presentation |
| 18 | 3/5/2020 | Final project | MSOBx303 | Final project presentation |
| 19 | 3/10/2020 | James | MSOBx303 | Zou Lab research presentation |
| 20 | 3/12/2020 | Manuel | MSOBx303 | Rivas Lab research presentation |
Assignments
Please submit all the assignments via Gradescope.
Late day policy. Students have 6 late days in total. We allow a maximum of 2 days per assignment.
Reading Materials
We ask students to write a paragraph about the reading materials and/or the corresponding lecture. Here is the instructions:
- Please read the reading materials posted on the class website and write a paragraph about it. You may write more if you want.
- You can write anything regarding the reading material and/or the corresponding lecture.
- You may also include comments and/or request regarding the teaching style. We appreciate your feedback!
- Please submit your answer through Gradescope as one pdf document.
Problem set 1
- Release date: 1/14/2020, Due date: 1/30/2020 (extended from 1/28/2020)
- Problem set 1
- Answer key
- Regrade request due: 2/25 (Tue.) 11:59 pm.
Problem set 2
- Release date: 1/30/2020, Due date: 2/18/2020
- Problem set 2
- Additional reference
- Answer key
- Sample code
- Regrade request due: 3/17 (Tue.) 11:59 pm.
Class project
We think the class project is a great opportunity for you to use some of the methods you will learn in the class. To allocate sufficient time to work on the project, we would like to have the brief project proposal by the third week of the quarter, 1/23/2020.
Project proposal
- Release date: 1/9/2020, Due date: 1/23/2020
Please check this document for more details.
Project write-up
- Due date: 3/12/2020
- There is no page limit, but We would like to see the following contents:
- Research question
- Motivation/Background
- Method
- Please clearly describe the statistical/deep learning model you’ve used in the class project
- Results & Discussion
- Make sure to include all the main findings from the analysis.
- For figures and tables, please use clear labels and provide legend so that we have a clear ideas what is presented on the figures/tables.
- For group project, please clarify the contribution of each group member in the final project write-up.
- If you are working in a lab, you may use some of the results from your research as the class project, given that PI(s) are aware and supportive about it. If you were to take this route, we will ask you to clarify the relevance of the project to the materials covered in the class.
Lecture materials
We will post the list of lecture slides and reading materials here.
Lecture 1. Overview of emerging topics in biomedical data science
- Lecture slides (on Canvas)
- D. Donoho. 50 years of Data Science
- Chapter 18 and Epilogue of Computer Age Statistical Inference
Lecture 2. Topics in Linear Algebra
- Lecture slides (on Canvas)
- Tibshirani, R. In praise of sparsity and convexity. in Past, Present, and Future of Statistical Science (eds. Lin, X. et al.) 497–505 (Chapman and Hall/CRC, 2014). doi:10.1201/b16720-47.
Lecture 3. Optimization
- Lecture slides (on Canvas)
- Constrained Optimization — Computational Statistics and Statistical Computing 1.0 documentation.
Lecture 4. Mixture Models
- Lecture slides (on Canvas)
- Rivas, M. A. et al. Effect of predicted protein-truncating genetic variants on the human transcriptome. Science 348, 666–669 (2015).
- Rivas, M. A. et al. Insights into the genetic epidemiology of Crohn’s and rare diseases in the Ashkenazi Jewish population. PLOS Genetics 14, e1007329 (2018).
- Flynn, E. et al. Sex-specific genetic effects across biomarkers. bioRxiv 837021 (2019) doi:10.1101/837021.
Lecture 5. Machine Learning for CRISPR editing
Lecture 6. LassoNet (guest lecture)
- Lecture slides (on Canvas)
- Lemhadri, I., Ruan, F. & Tibshirani, R. A neural network with feature sparsity. arXiv:1907.12207 [cs, stat] (2019).
Lecture 7. Causal inference using instrumental variables
- Lecture slides (on Canvas)
- Burgess, S., Foley, C. N. & Zuber, V. Inferring Causal Relationships Between Risk Factors and Outcomes from Genome-Wide Association Study Data. Annual Review of Genomics and Human Genetics 19, 303–327 (2018). (please clink “View on Journal Site”)
- Hernan, M. A. & Robins, J. M. Instruments for Causal Inference: An Epidemiologist??s Dream? Epidemiology 17, 360–372 (2006).
- P Cannon, C. IMPROVE-IT Trial: A Comparison of Ezetimibe/Simvastatin versus Simvastatin Monotherapy on Cardiovascular Outcomes After Acute Coronary Syndromes. in (2014).
- Burgess, S., Foley, C. N. & Zuber, V. Inferring Causal Relationships Between Risk Factors and Outcomes from Genome-Wide Association Study Data. Annual Review of Genomics and Human Genetics 19, 303–327 (2018).
Lecture 8. Causal inference using instrumental variables II: Mendelian randomization
- Lecture slides (on Canvas)
- Darrous, L., Mounier, N. & Kutalik, Z. Simultaneous estimation of bi-directional causal effects and heritable confounding from GWAS summary statistics. medRxiv 2020.01.27.20018929 (2020) doi:10.1101/2020.01.27.20018929.
- Burgess, S., Foley, C. N. & Zuber, V. Inferring Causal Relationships Between Risk Factors and Outcomes from Genome-Wide Association Study Data. Annual Review of Genomics and Human Genetics 19, 303–327 (2018).
Lecture 9. Gaussian Process regression
- Lecture slides (on Canvas)
- Chapter 2 Gaussian Process regression, Rasmussen, C. E. & Williams, C. K. I. Gaussian processes for machine learning. (MIT Press, 2006).
- 1.7. Gaussian Processes — scikit-learn 0.22.1 documentation.
- Bayesian non-parametrics with Gaussian Processes
Lecture 10. Reproducible large-scale inference
- Lecture slides (on Canvas)
- Ioannidis, J. P. A. . Why Most Published Research Findings Are False. PLoS Medicine 2, e124 (2005).
- Write Your Own R Packages.
- Coding habits for data scientists. ThoughtWorks (2019).
- Thomas, K. et al. Jupyter Notebooks - a publishing format for reproducible computational workflows. Stand Alone 87–90 (2016).
Lecture 11. Deep Learning I
Lecture 12. Deep Learning II
- Lecture slides (on Canvas)
- Zou, J. et al. A primer on deep learning in genomics. Nature Genetics (2018).
Lecture 13. Risk Models
- Lecture slides (on Canvas)
- Jostins, L. & Barrett, J. C. Genetic risk prediction in complex disease. Hum Mol Genet 20, R182–R188 (2011).
- Qian, J. et al. A Fast and Flexible Algorithm for Solving the Lasso in Large-scale and Ultrahigh-dimensional Problems. bioRxiv 630079 (2019).
Lecture 14. Survival risk models
- Lecture slides (on Canvas)
- Bewick, V., Cheek, L. & Ball, J. Statistics review 12: Survival analysis. Crit Care 8, 389 (2004).
- Li, R. et al. Fast Lasso method for Large-scale and Ultrahigh-dimensional Cox Model with applications to UK Biobank. bioRxiv 2020.01.20.913194 (2020).
Lecture 15. Multitask risk modeling
- Lecture slides (on Canvas)
- Tanigawa, Y.*, Li, J.* et al. Components of genetic associations across 2,138 phenotypes in the UK Biobank highlight adipocyte biology. Nat Commun 10, 1–14 (2019).
- Aguirre, M. et al. Polygenic risk modeling with latent trait-related genetic components. bioRxiv 808675 (2019) doi:10.1101/808675.
Lecture 16. False discovery rates
- Lecture slides (on Canvas)
- Stephens, M. False discovery rates: a new deal. Biostatistics 18, 275–294 (2017).
Lecture 17 & 18. Final project presentation
Lecture 19. Zou Lab research presentation
- James Zou’s research group
- Ouyang, D. et al. Interpretable AI for beat-to-beat cardiac function assessment. medRxiv 19012419 (2019).
- Ghorbani, A. & Zou, J. Neuron Shapley: Discovering the Responsible Neurons. arXiv:2002.09815 [cs, stat] (2020).
- Abid, A. et al. Gradio: Hassle-Free Sharing and Testing of ML Models in the Wild. arXiv:1906.02569 [cs, stat] (2019).